In the process of conducting genetic studies, we often encounter insufficient RNA samples, for example, for studying tiny anatomical oral tumors, even single-cell samples, and samples of specific gene mutations that are transcribed at very low levels in human cells. Of course, for the COVID-19 test, if the swabs are not in the right place or not enough times during sampling, the sample size will be very low, which is why the Commission of Health and Family Planning came out two days ago and passed the test, and if the nucleic acid sampler did not take six samples, you can report it.
The sensitivity of the reagent is important because we have this problem or that problem, so what can we do to improve the sensitivity of RT-PCR?
Before we discuss possible solutions, let’s mention two big complications with the situation we just mentioned.
First of all, we worry about RNA loss when we have only a few cell populations in our sample. If traditional separation and cleaning methods are used, such as column method or nucleic acid precipitation method, there is a high possibility that the few samples will be lost. One solution is to add a carrier molecule, such as tRNA, but even then, there is no guarantee that our recovery experiment is OK.
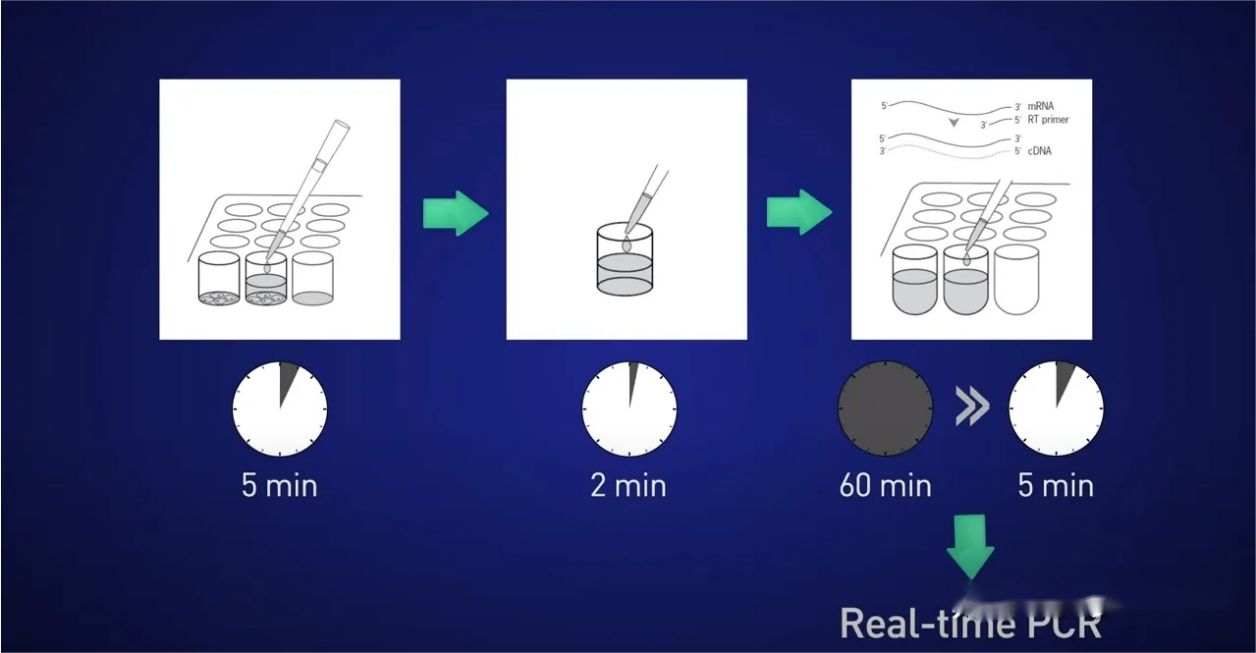
So what’s a better way? A good option for cultured cells or microanatomical samples is to use direct lysis.
The idea is to split the cells for 5 minutes, release the RNA into the solution, then stop the reaction for 2 minutes, then add the lysate directly to the reverse transcription reaction so that no RNA is lost, and finally put the resulting cDNA directly into the real-time reaction.
But what if, because of a limited starting point or a small amount of target gene expression, we can recycle all the RNA and still not provide enough templates to get a good real-time signal?
In this case, the pre-amplification step can be very useful.
The following is a scheme to increase sensitivity after reverse transcription. Before starting, we need to ask downstream which targets we are interested in, so as to design specific primers for these targets for pre-amplification.
This can be achieved by creating a mixed primer with up to 100 pairs of primers and a reaction cycle of 10 to 14 times. Therefore, a Master Mix specifically designed for this requirement is needed to pre-amplify the cDNA obtained.
The reason for setting the number of cycles between 10 and 14 is that this limited number of cycles ensures randomness between the various targets, which is crucial for researchers who need quantitative molecular information.
After pre-amplification, we can get a large amount of cDNA, so that the detection sensitivity at the back-end is greatly improved, and we can even dilute the sample and perform multiple real-time PCR reactions to eliminate possible random errors.
Post time: Apr-11-2023